Evolutionary Rates & Selection Mode (Mr.Bayes)
2025-08-27
Source:vignettes/rates-selection_MrBayes.Rmd
rates-selection_MrBayes.RmdThis vignette explains how to extract evolutionary rate parameters estimated from relaxed clock Bayesian inference analyses produced by Mr. Bayes. It also shows how to use evolutionary rate based inference of selection mode (strength) adapted to clock-based rates, as introduced by Simões and Pierce (2021). See the sister vignette “Evolutionary Rates & Selection Mode (BEAST2)” for an equivalent workflow using output data produced by BEAST2
Load the EvoPhylo package
Evolutionary Rates Statistics and Plots
In this section, we will extract evolutionary rate parameters from each node from a Bayesian clock (time-calibrated) summary tree. The functions below will store them in a data frame, produce summary statistics tables, and create different plots showing how rates are distributed across morphological partitions and clades.
1. Get rates from the clock tree and create a rate table
First, import a Bayesian clock tree using treeio’s
function read.mrbayes() (= read.beast()).
## Import summary tree with three clock partitions produced by
## Mr. Bayes (.t or .tre files) from your local directory
tree3p <- treeio::read.mrbayes("Tree3p.t")Below, we use the example Mr.Bayes single multi-clock tree with 3
morphological partitions tree3p from Simões and Pierce (2021) that accompanies
EvoPhylo.
data(tree3p)Subsequently, using get_clockrate_table_MrBayes(), users
can extract mean or median rate values for each node in the summary tree
that were annotated by Mr. Bayes when creating
the summary tree with Mr. Bayes “sumt”
command. These mean or median rate values are calculated by Mr. Bayes taking into
account all trees from the posterior sample. This works for any summary
tree produced by Mr. Bayes: a majority
rule consensus or the fully resolved maximum compatible tree (the latter
is used in the examples here).
Please note that analyses must have reached the stationarity phase and independent runs converging for the summary statistics in each node to be meaningful summaries of the posterior sample.
## Get table of clock rates with summary stats for each node in
## the tree for each relaxed clock partition (3 partitions in this tree file)
RateTable_Means_3p <- get_clockrate_table_MrBayes(tree3p, summary = "mean")
2. Export the rate table and plot tree with node values
Once a rate table has been obtained from Mr. Bayes it is necessary to export it. This is a necessary step to subsequently open the rate table spreadsheet locally (e.g., using Microsoft Office Excel) and customize the table with clade names associated with with each node in the tree for downstream analyses. Note that the root node may include “NA” for rate value, so it must be removed from the rate table.
## Export the rate tables (using Mr. Bayes example with 3 partitions)
write.csv(RateTable_Means_3p, file = "RateTable_Means_3p.csv")Plot tree node labels to customize clade names
To visualize the node values in the tree, you can use
ggtree().
## Plot tree node labels
library(ggtree)
tree_nodes <- ggtree(tree3p, branch.length = "none", size = 0.05) +
geom_tiplab(size = 2, linesize = 0.01, color = "black", offset = 0.5) +
geom_label(aes(label = node), size = 2, color="purple")
tree_nodes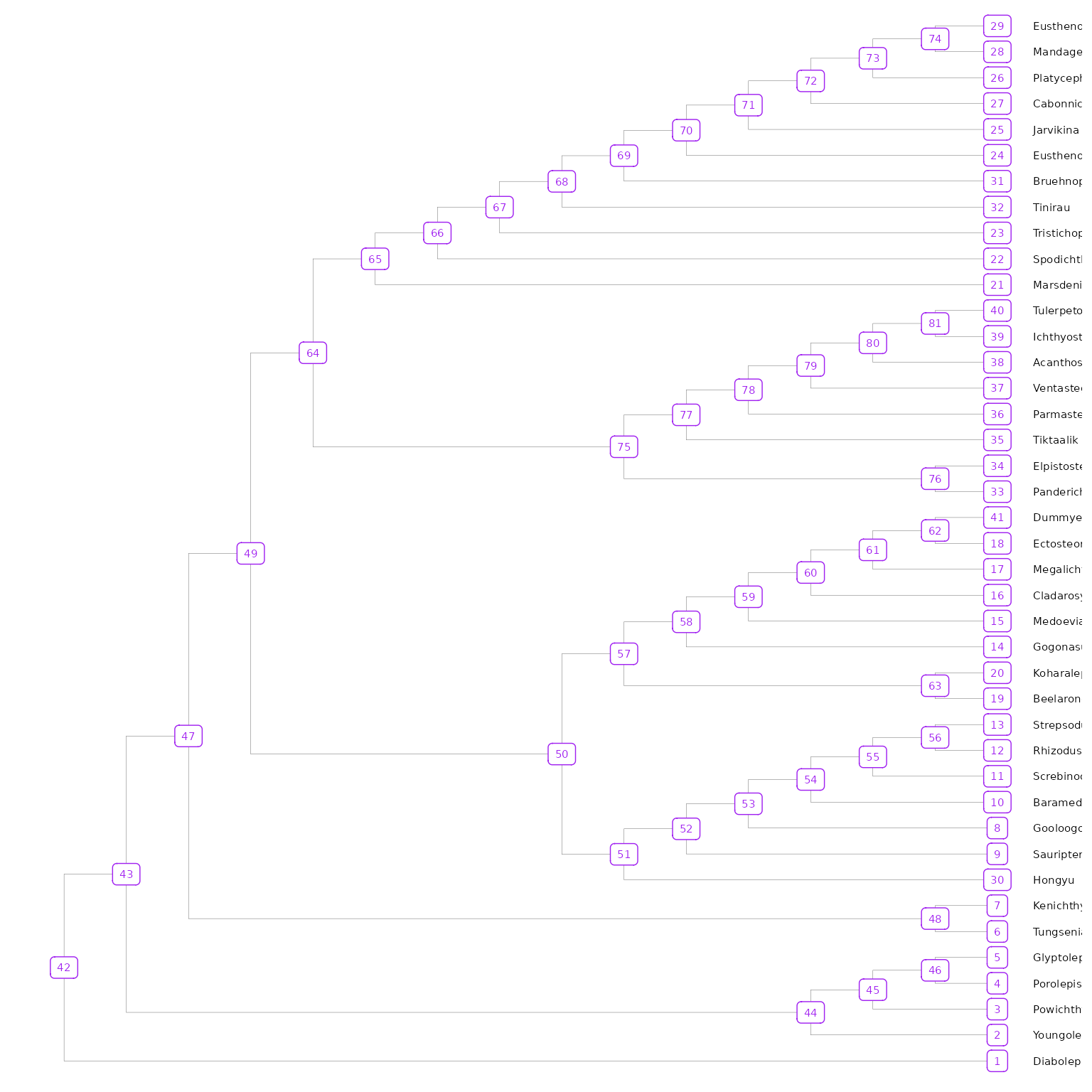
## Save your plot to your working directory as a PDF
ggplot2::ggsave("Tree_nodes.pdf", width = 10, height = 10)
3. Import rate table with custom clade memberships
A new “clade” column has been added to the rates table
## Import rate table with clade membership (new "clade" column added)
## from your local directory
RateTable_Means_3p_Clades <- read.csv("RateTable_Means_3p_Clades.csv", header = TRUE)Below, we use the rate table with clade membership
RateTable_Means_3p_Clades that accompanies
EvoPhylo.
data(RateTable_Means_3p_Clades)
head(RateTable_Means_3p_Clades)
## clade nodes rates1 rates2 rates3
## 1 Dipnomorpha 1 0.943696 0.981486 1.006164
## 2 Dipnomorpha 2 1.065326 0.772074 0.913194
## 3 Dipnomorpha 3 1.182460 0.656872 0.813618
## 4 Dipnomorpha 4 1.229767 0.523709 0.722519
## 5 Dipnomorpha 5 1.230564 0.517773 0.720479
## 6 Other 6 0.658855 0.717277 0.663950
4. Get summary stats for each clade/clock partition
Obtain summary statistics table and plots for each clade by clock
partition using clockrate_summary(). Supplying a file path
to file save the output to that file.
## Get summary statistics table for each clade by clock
clockrate_summary(RateTable_Means_3p_Clades,
file = "Sum_RateTable_Means_3p.csv")| clade | clock | n | mean | sd | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|---|
| Dipnomorpha | 1 | 8 | 1.10 | 0.11 | 0.94 | 1.02 | 1.10 | 1.19 | 1.23 |
| Elpisostegalia | 1 | 14 | 1.61 | 0.22 | 1.13 | 1.45 | 1.68 | 1.80 | 1.81 |
| Osteolepididae | 1 | 11 | 0.63 | 0.26 | 0.16 | 0.44 | 0.81 | 0.84 | 0.87 |
| Rhizodontidae | 1 | 14 | 0.57 | 0.30 | 0.03 | 0.33 | 0.67 | 0.83 | 0.89 |
| Tristichopteridae | 1 | 21 | 0.71 | 0.04 | 0.61 | 0.69 | 0.72 | 0.74 | 0.78 |
| Other | 1 | 11 | 0.89 | 0.36 | 0.54 | 0.69 | 0.78 | 0.94 | 1.81 |
| Dipnomorpha | 2 | 8 | 0.75 | 0.18 | 0.52 | 0.62 | 0.75 | 0.89 | 0.98 |
| Elpisostegalia | 2 | 14 | 1.36 | 0.10 | 1.03 | 1.36 | 1.38 | 1.41 | 1.42 |
| Osteolepididae | 2 | 11 | 0.34 | 0.15 | 0.07 | 0.28 | 0.38 | 0.45 | 0.53 |
| Rhizodontidae | 2 | 14 | 0.33 | 0.18 | 0.02 | 0.17 | 0.38 | 0.44 | 0.56 |
| Tristichopteridae | 2 | 21 | 0.34 | 0.06 | 0.27 | 0.32 | 0.33 | 0.33 | 0.55 |
| Other | 2 | 11 | 0.75 | 0.25 | 0.39 | 0.61 | 0.72 | 0.78 | 1.35 |
| Dipnomorpha | 3 | 8 | 0.87 | 0.11 | 0.72 | 0.79 | 0.89 | 0.95 | 1.01 |
| Elpisostegalia | 3 | 14 | 0.83 | 0.16 | 0.63 | 0.67 | 0.89 | 0.99 | 1.00 |
| Osteolepididae | 3 | 11 | 0.32 | 0.13 | 0.07 | 0.27 | 0.33 | 0.42 | 0.49 |
| Rhizodontidae | 3 | 14 | 0.32 | 0.17 | 0.02 | 0.21 | 0.40 | 0.43 | 0.52 |
| Tristichopteridae | 3 | 21 | 0.52 | 0.08 | 0.37 | 0.44 | 0.54 | 0.59 | 0.64 |
| Other | 3 | 11 | 0.73 | 0.17 | 0.47 | 0.64 | 0.70 | 0.81 | 1.00 |
5. Plot rates by clock partition and clade
Plot distributions of rates by clock partition and clade with
clockrate_dens_plot().
## Overlapping plots
clockrate_dens_plot(RateTable_Means_3p_Clades, stack = FALSE,
nrow = 1, scales = "fixed")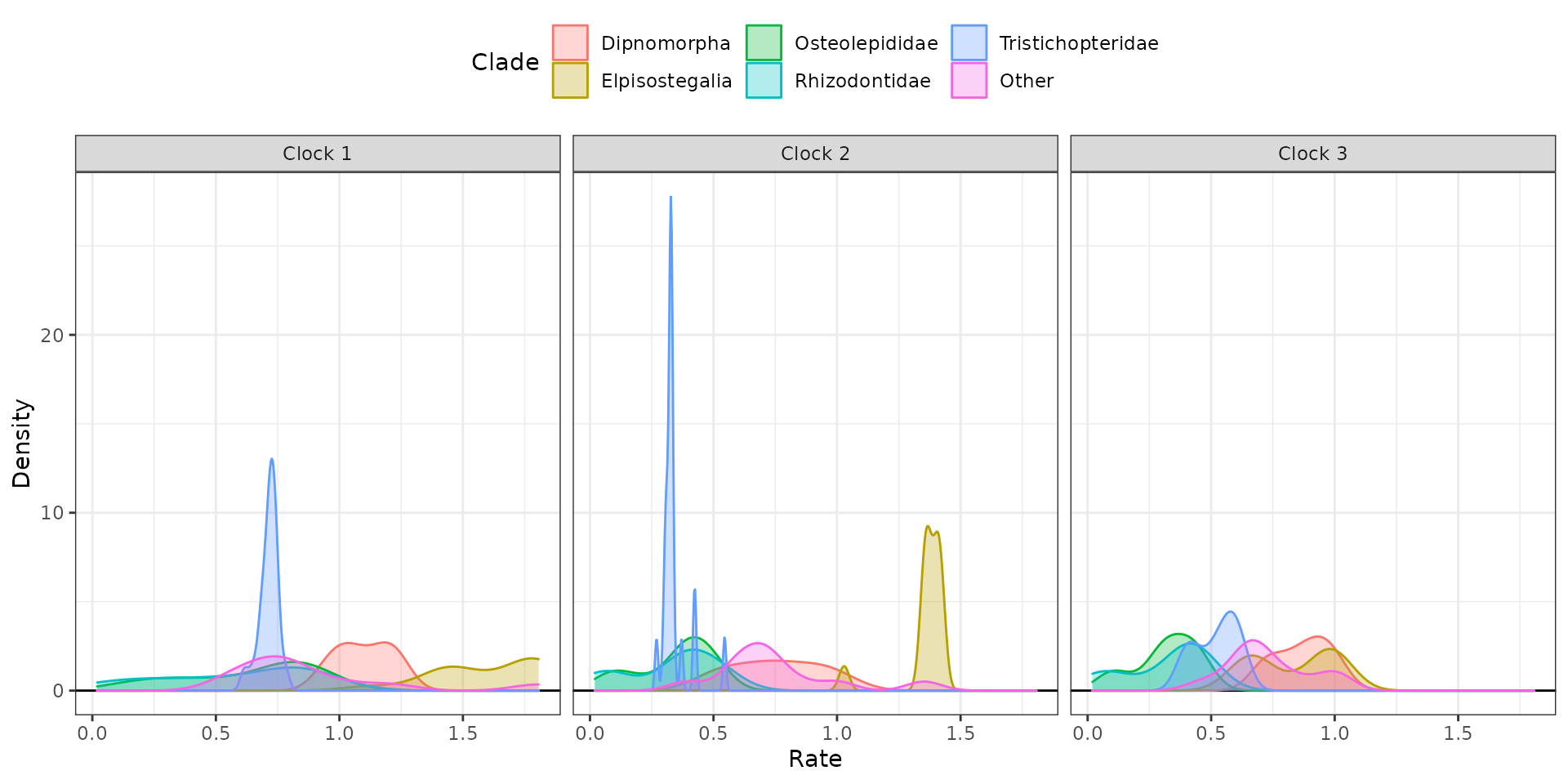
Sometimes using stacked plots provides a better visualization as it avoids overlapping distributions.
## Stacked plots
clockrate_dens_plot(RateTable_Means_3p_Clades, stack = TRUE,
nrow = 1, scales = "fixed")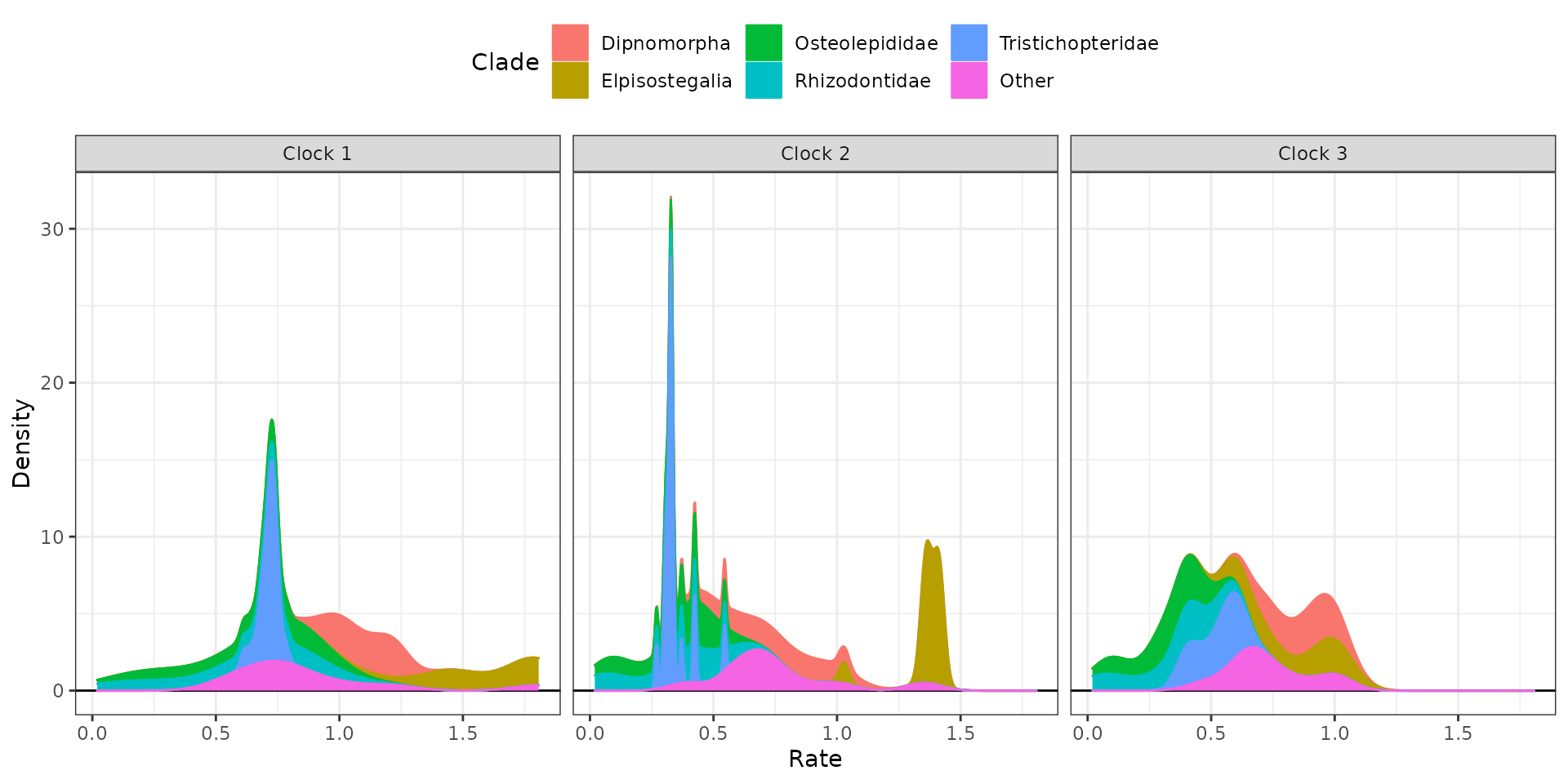
It is also possible to append extra layers using ggplot2
function, such as for changing the color scale. Below, we change the
color scale to be the Viridis scale.
## Stacked plots with viridis color scale
clockrate_dens_plot(RateTable_Means_3p_Clades, stack = TRUE,
nrow = 1, scales = "fixed") +
ggplot2::scale_color_viridis_d() +
ggplot2::scale_fill_viridis_d()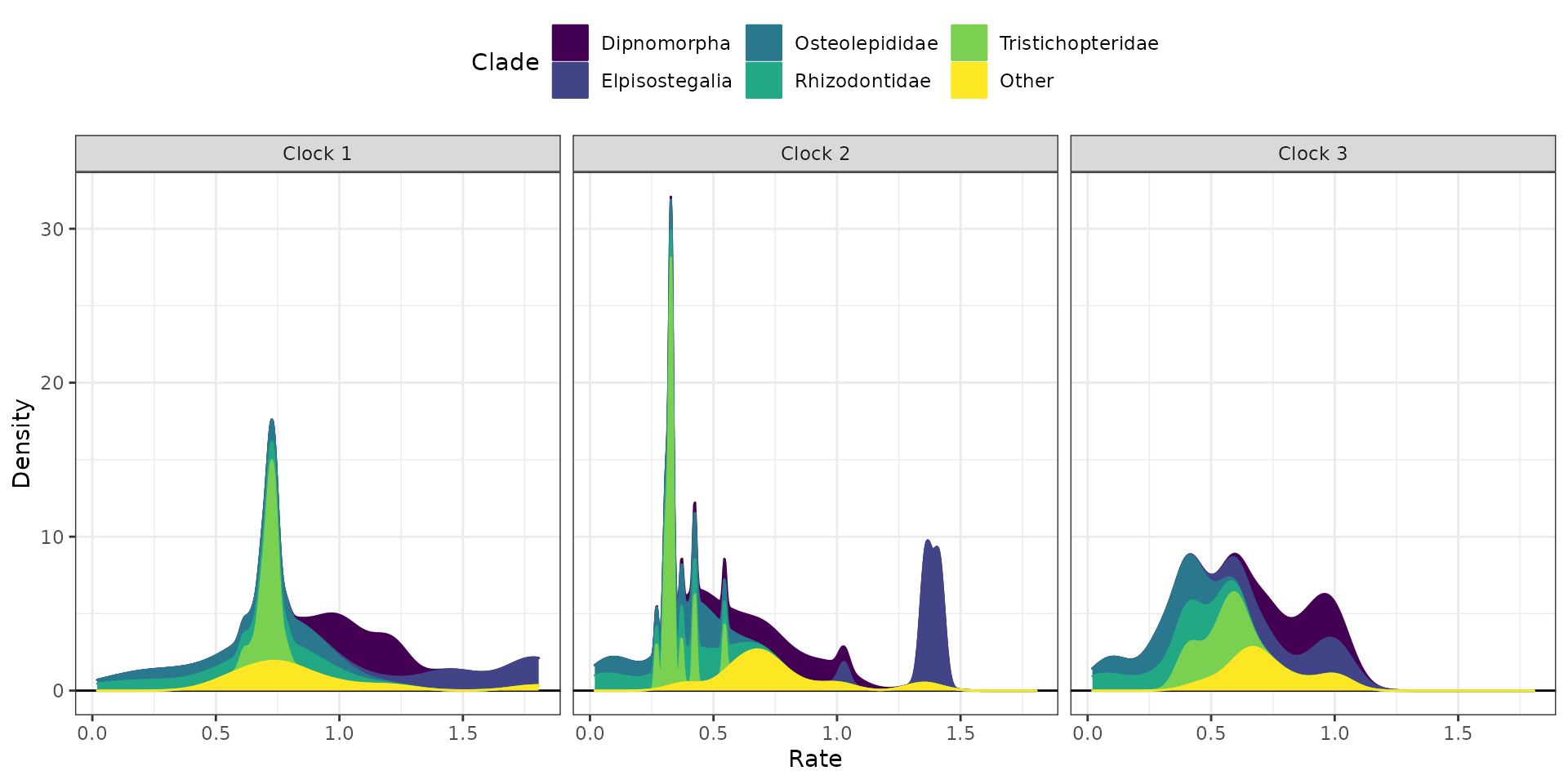
6. Rate linear models
We can also plot linear model regressions between rates from two or
more clocks with clockrate_reg_plot().
## Plot regressions of rates from two clocks
p12 <- clockrate_reg_plot(RateTable_Means_3p_Clades, clock_x = 1, clock_y = 2)
p13 <- clockrate_reg_plot(RateTable_Means_3p_Clades, clock_x = 1, clock_y = 3)
p23 <- clockrate_reg_plot(RateTable_Means_3p_Clades, clock_x = 2, clock_y = 3)
library(patchwork) #for combining plots
p12 + p13 + p23 + plot_layout(ncol = 2)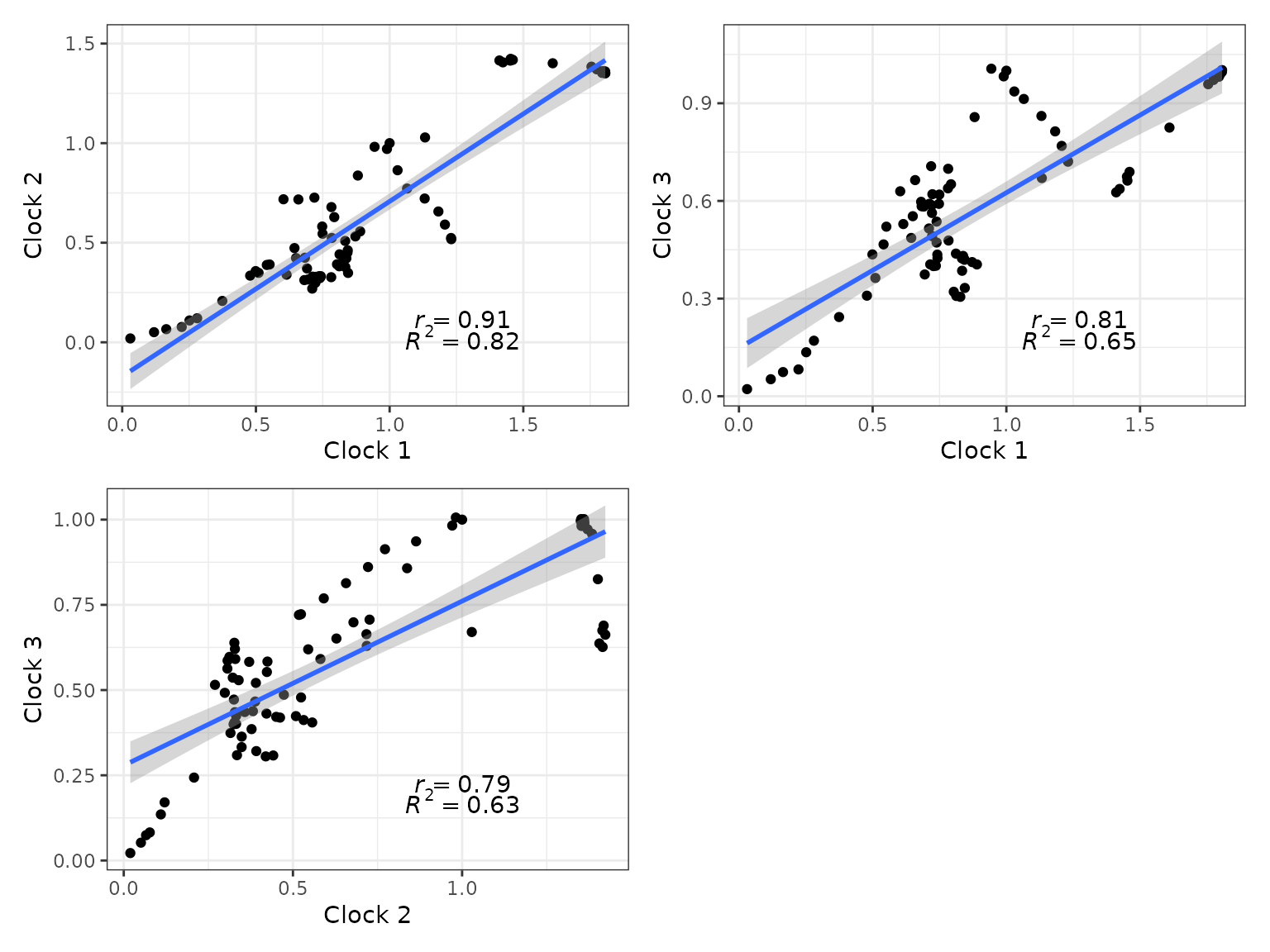
## Save your plot to your working directory as a PDF
ggplot2::ggsave("Plot_regs.pdf", width = 8, height = 8)
ADDENDUM: Example using rates from single clock analysis (no partitioning)
You can also explore clock rates for summary trees including a single clock shared among all character partitions (or an unpartitioned analysis):
## Import summary tree with a single clock partitions produced by
## Mr. Bayes (.t or .tre files) from examples directory
tree1p <- treeio::read.mrbayes("Tree1p.t")Below, we use the example tree tree1p that accompanies
EvoPhylo.
data(tree1p)Then, get table of clock rates with summary stats for each node in the tree for each relaxed clock partition.
RateTable_Means_1p <- get_clockrate_table_MrBayes(tree1p, summary = "mean")Then proceed with the analysis, as in the case with multiple clocks.
## Export the rate tables
write.csv(RateTable_Means_1p, file = "RateTable_Means1.csv")
## Import rate table after adding clade membership (new "clade" column added)
RateTable_Means_1p_Clades <- read.csv("RateTable_Means1_Clades.csv", header = TRUE)
#Below, we use the rate table with clade membership `RateTable_Means_1p_Clades` that accompanies `EvoPhylo`.
data(RateTable_Means_1p_Clades)
## Get summary statistics table for each clade by clock
clockrate_summary(RateTable_Means_1p_Clades,
file = "Sum_RateTable_Medians1.csv")| clade | n | mean | sd | min | Q1 | median | Q3 | max |
|---|---|---|---|---|---|---|---|---|
| Dipnomorpha | 8 | 0.57 | 0.28 | 0.22 | 0.37 | 0.54 | 0.78 | 0.95 |
| Elpisostegalia | 14 | 0.91 | 0.25 | 0.44 | 0.77 | 0.85 | 1.03 | 1.35 |
| Osteolepididae | 11 | 0.23 | 0.10 | 0.03 | 0.18 | 0.23 | 0.30 | 0.38 |
| Rhizodontidae | 14 | 0.18 | 0.15 | 0.00 | 0.04 | 0.20 | 0.29 | 0.42 |
| Tristichopteridae | 21 | 0.39 | 0.43 | 0.05 | 0.11 | 0.19 | 0.34 | 1.32 |
| Other | 11 | 0.41 | 0.26 | 0.20 | 0.25 | 0.28 | 0.45 | 1.00 |
## Stacked plots with viridis color scale
clockrate_dens_plot(RateTable_Means_1p_Clades, stack = TRUE,
nrow = 1, scales = "fixed") +
ggplot2::scale_color_viridis_d() +
ggplot2::scale_fill_viridis_d()
Selection Mode
In this section, we will use evolutionary rate based inference of selection mode, as first introduced by Baker et al. (2016) for continuous traits, and later adapted to clock-based rates by Simões and Pierce (2021).
1. Import combined log file from all runs.
This is produced by using combine_log(). The first
argument passed to combine_log() should be a path to the
folder containing the log files to be imported and combined.
## Import all log (.p) files from all runs and combine them, with burn-in = 25%
## and downsampling to 2.5k trees in each log file
posterior3p <- combine_log("LogFiles3p", burnin = 0.25, downsample = 1000)Below, we use the posterior dataset posterior3p that
accompanies EvoPhylo.
###2. Check background rates distribution and if they need transformation
The output includes histograms showing the data distribution and mean (red dotted line) before and after data transformation for comparisons.
library(ggplot2)
B <- plot_back_rates (type = "MrBayes", posterior3p, clock = 1,
trans = "log10", size = 10, quantile = 1)
B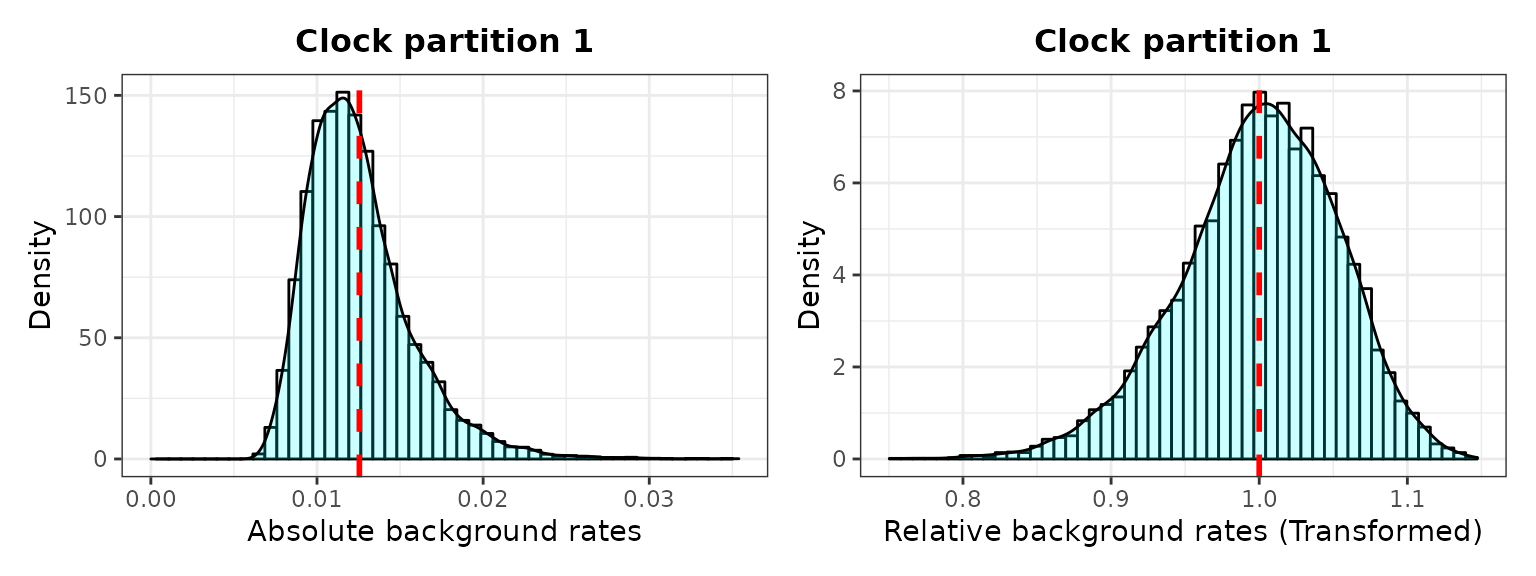
3. Plot selection gradient on the summary tree
Using different thresholds, identify the selection mode across
branches in the tree for each clock partition with
plot_treerates_sgn().
Users must indicate the type of output file (between Mr. Bayes and BEAST2) and whether they would like
to log transform the background rate to meet assumptions of normally
distributed data, based on the results obtained from
plot_back_rates. Users should also indicate in “clock” the
number of the clock partition they would like to plot rates from and the
desired significance threshold to interpret branch rates (we recommend
number of standard deviations around the mean of background
rates).Finally, a series of arguments enable users to customize the
geological timescale to add to the tree.
## Plot tree using various thresholds for clock partition 1
A1 <- plot_treerates_sgn(
type = "MrBayes", trans = "none", #Indicates software name output and type of transformation
tree3p, posterior3p, #Summary tree and posterior files
clock = 1, #Show rates for clock partition 1
summary = "mean", #sets summary stats to get from summary tree nodes
branch_size = 1.5, tip_size = 3, #sets size for tree elements
xlim = c(-450, -260), nbreaks = 8, geo_size = list(3, 3), #sets limits and breaks for geoscale
threshold = c("1 SD", "3 SD")) #sets threshold for selection mode
A1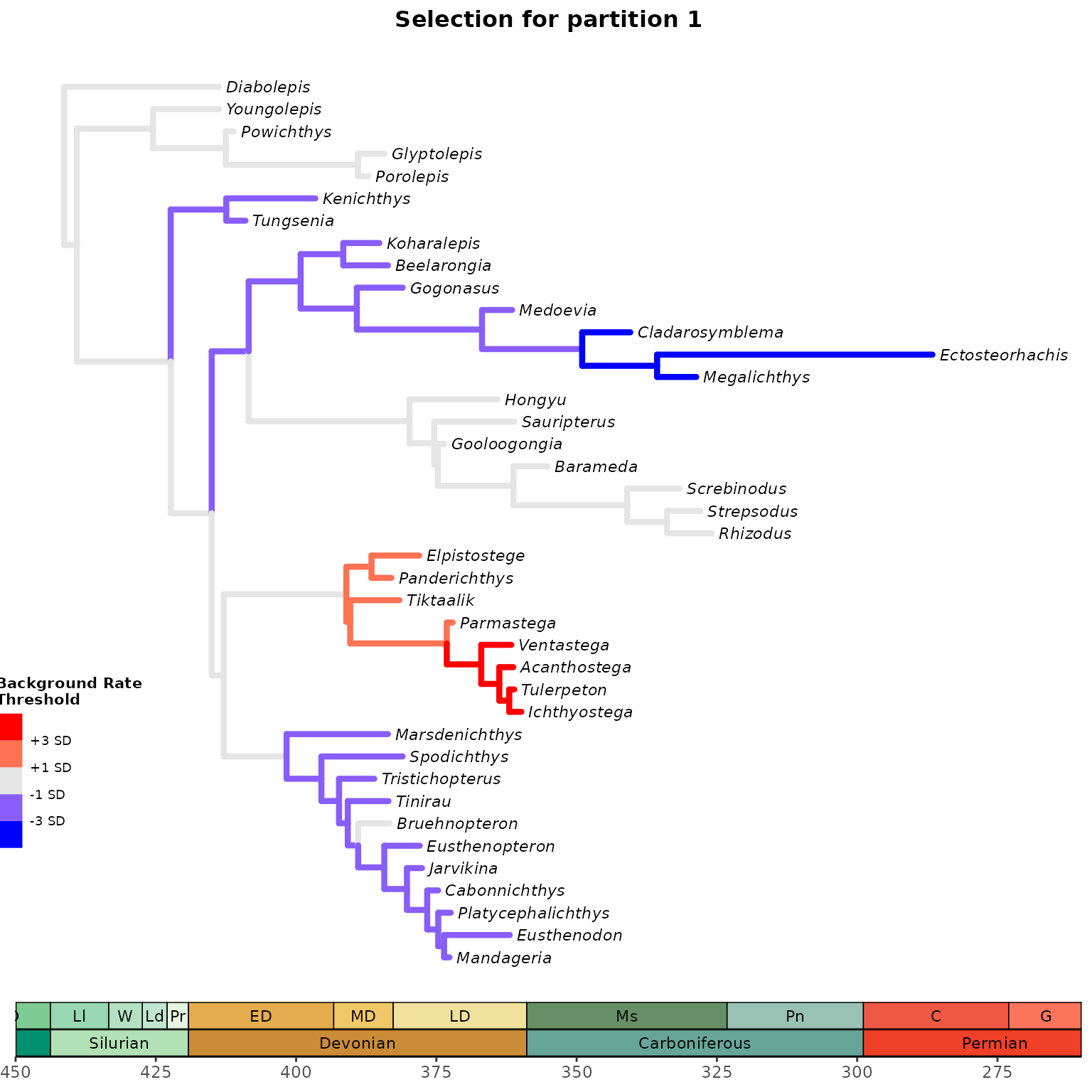
Plot tree using various thresholds for the other clock partitions and combine them.
## Plot tree using various thresholds for other clock partition and combine them
A2 <- plot_treerates_sgn(
type = "MrBayes", trans = "none", #Indicates software name output and type of transformation
tree3p, posterior3p, #Summary tree and posterior files
clock = 2, #Show rates for clock partition 1
summary = "mean", #sets summary stats to get from summary tree nodes
branch_size = 1.5, tip_size = 3, #sets size for tree elements
xlim = c(-450, -260), nbreaks = 8, geo_size = list(3, 3), #sets limits and breaks for geoscale
threshold = c("1 SD", "3 SD")) #sets threshold for selection mode
A3 <- plot_treerates_sgn(
type = "MrBayes", trans = "none", #Indicates software name output and type of transformation
tree3p, posterior3p, #Summary tree and posterior files
clock = 3, #Show rates for clock partition 1
summary = "mean", #sets summary stats to get from summary tree nodes
branch_size = 1.5, tip_size = 3, #sets size for tree elements
xlim = c(-450, -260), nbreaks = 8, geo_size = list(3, 3), #sets limits and breaks for geoscale
threshold = c("1 SD", "3 SD")) #sets threshold for selection mode
library(patchwork)
A1 + A2 + A3 + plot_layout(nrow = 1)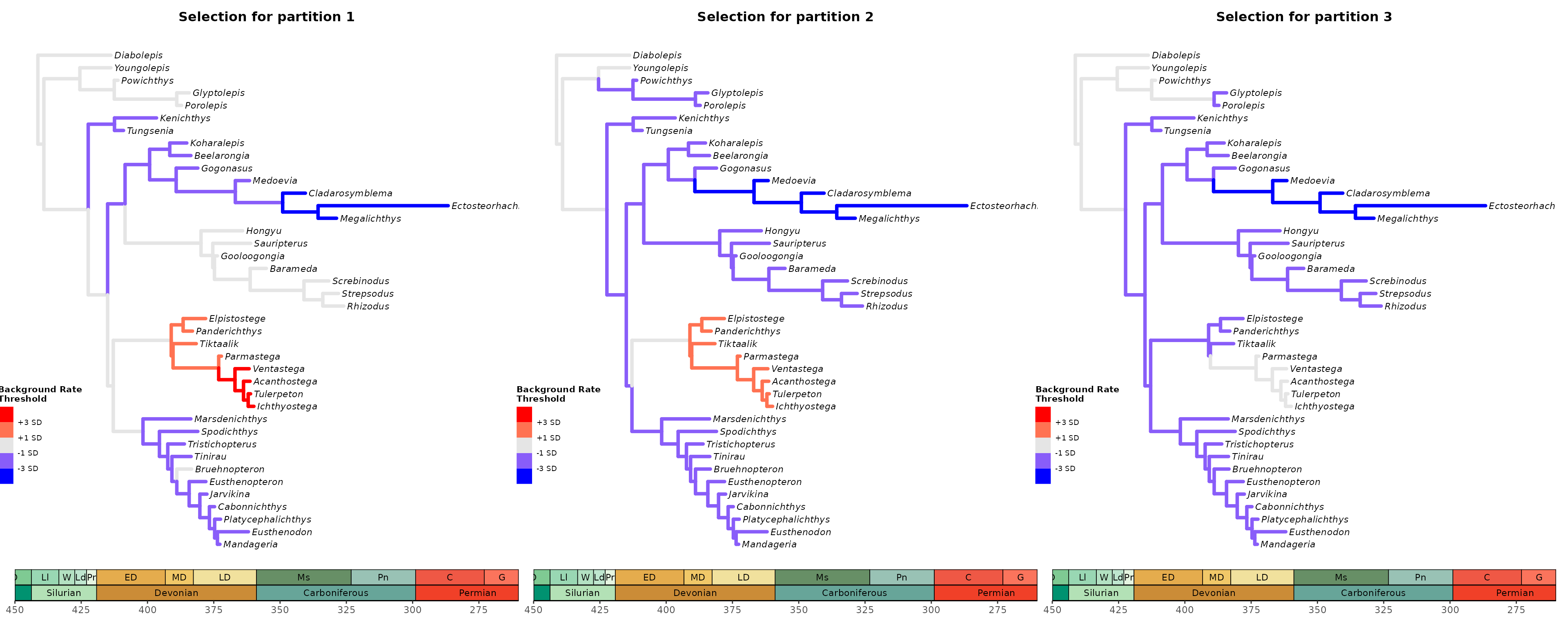
## Save your plot to your working directory as a PDF
ggplot2::ggsave("Tree_Sel_3p.pdf", width = 20, height = 8)
4. Pairwise t-tests of rate values among clades
The function get_pwt_rates_MrBayes() complements the
functionality of plot_treerates_sgn by producing a table of
pairwise t-tests for differences between the mean background rate in the
posterior and the absolute rate for each summary tree branches Should be
used only for normally distributed data in which a CI=0.95 is considered
a good threshold. In many cases, however, using multiple standard
deviations as outputted using plot_treerates_sgn provides a
more robust test of whether branch rates are significantly different
from background rates.
4.1. Import rate table with clade membership (new “clade” column added) from your local directory with “mean” values
## Import rate table with clade membership
RateTable_Means_3p_Clades <- read.csv("RateTable_Means_Clades.csv", header = TRUE)Below, we use the rate table with clade membership
RateTable_Means_3p_Clades that accompanies
EvoPhylo.
data(RateTable_Means_3p_Clades)4.2. Get and export table of pairwise t-tests
## Get table of pairwise t-tests for difference between the posterior
## mean and the rate for each tree node
RateSign_Tests <- get_pwt_rates_MrBayes(RateTable_Means_3p_Clades, posterior3p)
## Show first 10 lines of table
head(RateSign_Tests, 10)| clade | nodes | clock | relative.rate.mean | absolute.rate.mean | p.value |
|---|---|---|---|---|---|
| Dipnomorpha | 1 | 1 | 0.943696 | 0.0118443 | 0 |
| Dipnomorpha | 2 | 1 | 1.065326 | 0.0133709 | 0 |
| Dipnomorpha | 3 | 1 | 1.182460 | 0.0148411 | 0 |
| Dipnomorpha | 4 | 1 | 1.229767 | 0.0154348 | 0 |
| Dipnomorpha | 5 | 1 | 1.230564 | 0.0154448 | 0 |
| Other | 6 | 1 | 0.658855 | 0.0082693 | 0 |
| Other | 7 | 1 | 0.603090 | 0.0075694 | 0 |
| Osteolepididae | 8 | 1 | 0.843373 | 0.0105852 | 0 |
| Osteolepididae | 9 | 1 | 0.872012 | 0.0109446 | 0 |
| Osteolepididae | 10 | 1 | 0.811473 | 0.0101848 | 0 |
## Export the table
write.csv(RateSign_Tests, file = "RateSign_Tests.csv")